MAIN PAGE
> Back to contents
Cybernetics and programming
Reference:
Kazakov M.A.
Automation of genetic similarity and genetic distance calculation using Nei criterion
// Cybernetics and programming.
2015. № 6.
P. 1-5.
DOI: 10.7256/2306-4196.2015.6.16744 URL: https://en.nbpublish.com/library_read_article.php?id=16744
Automation of genetic similarity and genetic distance calculation using Nei criterion
Kazakov Mukhamed Anatol'evich
junior researcher, Institute of Applied Mathematics and Automation
360000, Russia, respublika Kaardino-Balkariya, g. Nal'chik, ul. Ingushskaya, 9, kv. 48

|
f_wolfgang@mail.ru
|
|
 |
|
DOI: 10.7256/2306-4196.2015.6.16744
Received:
23-10-2015
Published:
19-01-2016
Abstract:
The subject of the study is the problem of automation of genetic similarity and genetic distance calculation and building phylogenetic trees. The object of the research is the sequence of drives (alleles) in the arms of polytene chromosomes. The author considers in detail the chromosomal characteristics of species with seven chromosome arms by the example of chironomid mosquito (genus Chironomus). This article presents a Chironomus 1.0 software solution with a simple, intuitive user interface allowing to easily calculate the distance and genetic similarity between populations. The software is developed using C# and implements Nei method to calculate the genetic similarity and genetic distance. The special contribution made by the author in the study of the theme is in significant simplification of genetic studies relating to the calculation of genetic similarity and genetic distance using Nei method. The novelty of the research is in creation of a software product that allows to easily calculate the genetic similarity and distance for all types of populations with seven chromosome arms.
Keywords:
genetic distance, genetic similarity, computing automation, programs, Nei method, genetics, phylogenetic trees, chironomids, polytene chromosomes, software
This article written in Russian. You can find original text of the article here
.
Введение В данной статье представлена программа позволяющая вывчислять генетичесое сходство и генетическую дистанцию по кртитерию Нея между поппуляциями видов комаров комаров-звонцов рода Chironomus (Chironomidae, Diptera), а так же для любых других видов, имеющих семь плеч политенных хромосом. Метод определения генетического расстояния и генетического сходства Нея [1] основан на предположении того, что генетическая разница возникает за счет мутаций и генетического дрейфа.
На практике для расчетов генетического расстояния у Chironomus определяют частоты встречаемости в каждой популяции различных последовательностей дисков (аллелей) в семи плечах политенных хромосом (A, B, C, D, E, F и G). Представим себе, что для хромосомного плеча F существует i различных аллелей (1, 2, 3 и т.д.). В популяции А эти аллели встречаются с частотами ai; в популяции В частотам аллелей соответствуют bi. Исходя из этого, можно вычислить степень генетического сходства данных популяций по данному плечу:
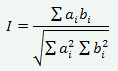
Само же генетическое расстояние D вычисляется по формуле` `
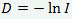
Значения генетического сходства могут варьировать от 0 (между популяциями нет общих аллелей), до 1 (частоты аллелей в популяции одинаковые), соответственно генетическая дистанция варьирует от 0 до ∞.
Автоматизация процесса вычисления генетического сходства и генетической дистанции для популяций рода Chironomus представляет практичесий интерес для задач построения филогенетических деревьев.
Существует несколько программных продуктов в которых предусмотрена возможноссть попарно вычислять цитогенетическое расстояние и цитогенетическое сходство по критерию Нея. Наиболее популярные из них, это пакеты программ PHYLIP [2] и MEGA 6 [3,4,5]. В первую очередь данные продукты предназначены для работы с нуклеотидными последовательностями ДНК и последовательностями аминокислот белков.
Вычисление генетических расстояний по критерию Нея у рода Chironomus возможно с помощью выше указанных пакетов программ. Однако в силу сложности процесса предварительной подготовки исходных данных для ввода и сложного, не интуитивного интерфейса, рассмотренные выше программные продукты не являются достаточно удобными вычислительными средствами для решения поставленной задачи. В силу чего возникает потребность в создании программного продукта, отвечающего следующим критериям:
- простота ввода исходных данных не требующей предварительной многошаговой обработки;
- интерфейс программы должен быть достаточно простым и интуитивно понятным;
- наглядность получаемых в результате вычислений;
- повышенная точность вычислений;
- возможность при необходимости добавления дополнительных методов вычислении генетической дистанции и генетического сходства;
- возможность одновременного сравнения большого числа популяций.
Результаты исследования В результате исследования был создан специализированный программный продукт Chironomus 1.0 (рис. 1), позволяющий быстро и наглядно вычислять генетические расстояния и генетическое сходство у рода Chironomus по методу Неи. Программа имеет простой русскоязычный интерфейс и позволяет одновременно сравнивать до 14 различных популяций, написана на языке программирования С# и имеет размер 120 Кбайт. Программа содержит 46 циклов и работает как приложение для платформы .NET Framework.
Процесс ввода данных и получение результата состоит из следующих шагов:
- Вводится количество сравниваемых популяций и число аллелей для каждого хромосомного плеча. Эти значения является первичными входными данными алгоритма программы;
- В соответствии с введенными значениями создается наглядная таблица для ввода частот аллелей популяций. Заполнение этой таблицы определяет вторичные входные данные алгоритма программы;
- После заполнения всей таблицы производится вычисление генетического сходства и генетической дистанции для каждой пары популяций, после чего производится автоматический вывод в поле результатов.
Программа была протестирована на данных по шести реально существующим популяциям вида Ch. bernensis Центрального Кавказе и Предкавказья [6]. Независимые расчеты генетических расстояний и генетических дистанции по тем же исходным данным подтвердили точность вычислений требуемых значений созданным нами программным продуктом.
References
1. Nei, M. The genetic distance between populations / M. Nei // American Naturalist. – 1972. – Vol. 106. – P. 283–29.
2. Felsenstein, J. Phylogeny Inference Package (PHYLIP) / J. Felsenstein // Version 3.5. – 1993. – University of Washington, Seattle.
3. Tamura, K. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods / K. Tamura, D. Peterson, N. Peterson, G. Stecher // Molecular Biology and Evolution – 2011. – Vol. 28. – P. 2731–2739.
4. Tamura, K. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0 / K. Tamura, G. Stecher, D. Peterson, A. Filipski // Molecular Biology and Evolution – 2013. –Vol. 30. – P. 2725 – 2729.
5. Kumar, S. MEGA-CC: computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis / S. Kumar, G. Stecher, D. Peterson, K. Tamura // Bioinformatics – 2012. – Vol. 28. – P. 2685 – 2686.
6. Karmokov, M.Kh. Karyotype characteristics and polymorphism peculiarities of Chironomus bernensis Wülker & Klötzli, 1973 (Diptera, Chironomidae) from the Central Caucasus and Ciscaucasia / M.Kh. Karmokov, N.V. Polukonova, O.V. Sinichkina // Comparative Cytogenetics. – 2015. Vol. 9(3). P. 281 – 297. doi: 10.3897/ CompCytogen.v9i3.451
Link to this article
You can simply select and copy link from below text field.
|





